Kishan K C
Applied Scientist
Amazon Alexa AI Search
About me
I am an Applied Scientist at Amazon Alexa AI Search, working on Question Answering systems. I joined Amazon as a full-time Applied Scientist in February 2022. During my internship at the Alexa Speaker identification team last summer, I worked with Zhenning Tan to develop an open-set few-shot learning framework to improve speaker identification.
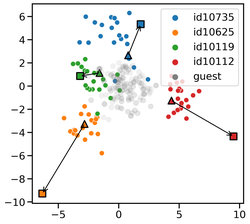
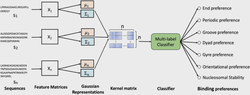
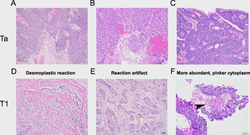
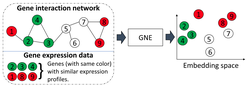
I obtained my Ph.D. in Computing and Information Sciences at Rochester Institute of Technology where I worked with Dr. Rui Li and Dr. Anne Haake. In my Ph.D. thesis, I focused on developing scalable probabilistic machine learning models to learn the representation of network structured data. Furthermore, I developed Bayesian model selection to infer the most plausible neural network structure warranted by data.
Interests
- Probabilistic Machine Learning
- Representation Learning
- Natural Languge Processing
- Link Prediction
Education
-
PhD in Computing and Information Sciences, 2022
Rochester Institute of Technology
-
B.E. in Computer Engineering, 2014
Institute of Engineering, Tribhuvan University